MATLAB and R
Many users find that they can only go so far when processing data with PAMGuard and there are often important steps that people need to take to go from PAMGuard to a published paper.
We therefore provide export functions and software libraries written in MATLAB and R (a Python version is on it’s way) which can access PAMGuard data files.
This allows you to conduct bespoke analysis of PAMGuard output in a more user friendly programming environment.
Export data to other platforms
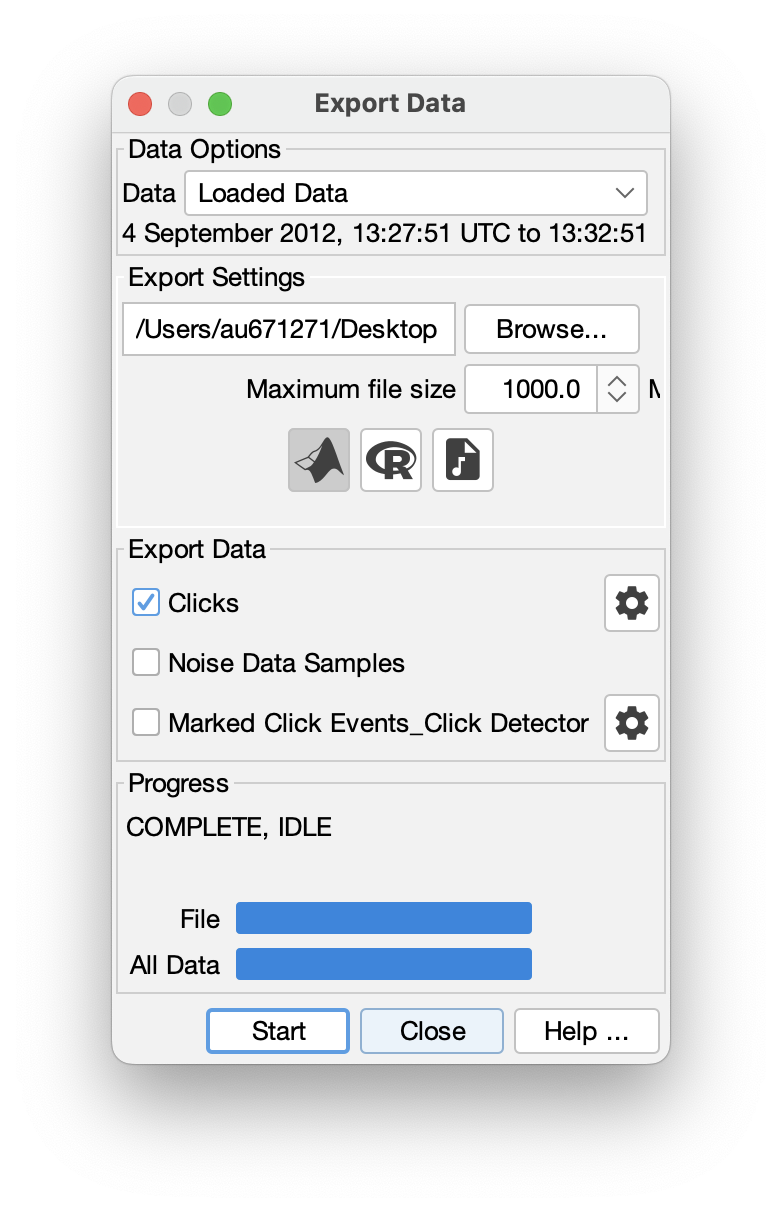
PAMGuard has a powerful exporter which can export detections to;
- .mat files which can be opened in MATLAB.
- .RData frames which can be open in R.
- .wav file which can be opened in any acoustics program.
The exporter allows filtering of data based on, for example, species classiifcaton, manual annotations etc. allowing users to specify exactly what is exported.
Thanks so much to our friends at NOAA Southwest Fisheries Science Center in San Diego , there is also an R package for opening PAMGuard binary files:
install.packages(“PamBinaries”)
You can find the source files on GitHub.
The best way to download and stay up-to-date with this is by following the installation instructions on the GitHub page. The R functions are written to work just like the MATLAB code, and there is a short tutorial on the GitHub page.
library(PamBinaries)
#count the number of classified clicks in a folder of pamguard files.
#folder to a set of binary files.
folder='my/folder/of/pamguard/click/files'
# list all the file names of the
# specified pattern
fnames <- list.files(folder, pattern = "Click_Detector_Clicks.*\\.pgdf$")
count =0; #the classified click counter.
for (val in fnames) {
print(val)
#load each click file.
clickfile <- loadPamguardBinaryFile(file.path(folder, val))
#iterate through the click files to count the classified clicks.
for (aclick in clickfile$data) {
if (aclick$type==1){
count=count+1
}
}
}Users can also take more control and import PAMGuard data files directly in MATLAB using the PAMGuard-matlab library
MATLAB code for opening PAMGuard binary files is now on the PAMGuard GitHub repository.
Clicking the green ‘Code’ button will allow you to download the code as a zip archive. Alternatively, you can clone the repository, which will make it easier to get updates if the code is updated.
% count the number of classified clicks in a binary file folder of clicks.
clear
% the path to the binary file foldeer
folder='/my/binary/file/folder';
binaryFiles = dir(fullfile(folder, 'Click_Detector_Clicks*.pgdf'));
count=0;
for i=1:length(binaryFiles)
%print progress
disp(['Loading files: ' num2str(i) ...
' of ' num2str(length(binaryFiles))])
%create filepath from dir dtruct
filePath=fullfile(folder, binaryFiles(i).name);
%load the clicks
clicks=loadPamguardBinaryFile(filePath);
%iterate through all clicks and count how many are classified
for j=1:length(clicks)
if (clicks(j).type==1)
count=count+1;
end
end
end