Data Export
The PAMGuard exporter allows users to export PAMGuard data, such as detections, to a variety of different formats. The exporter is a convenient solution for exporting sections or large chunks of a PAMGuard datasets without requiring any code. For more bespoke data management please see the PAMGuard-MATLAB library and PAMBinaries package which can be used for more bespoke data management. Note that the exporter only exports a sub set of data types - this will expand in future releases.
Exporting
The PAMGuard exporter can be accessed from File->Export. This brings up the Export dialog. The export dialog allows users to select which data to export, where to export it and the file format to export as. Each data block also has a settings icon which opens the data block’s unique data selector. So for example, users can export only specific types of clicks or whistles between certain frequencies.
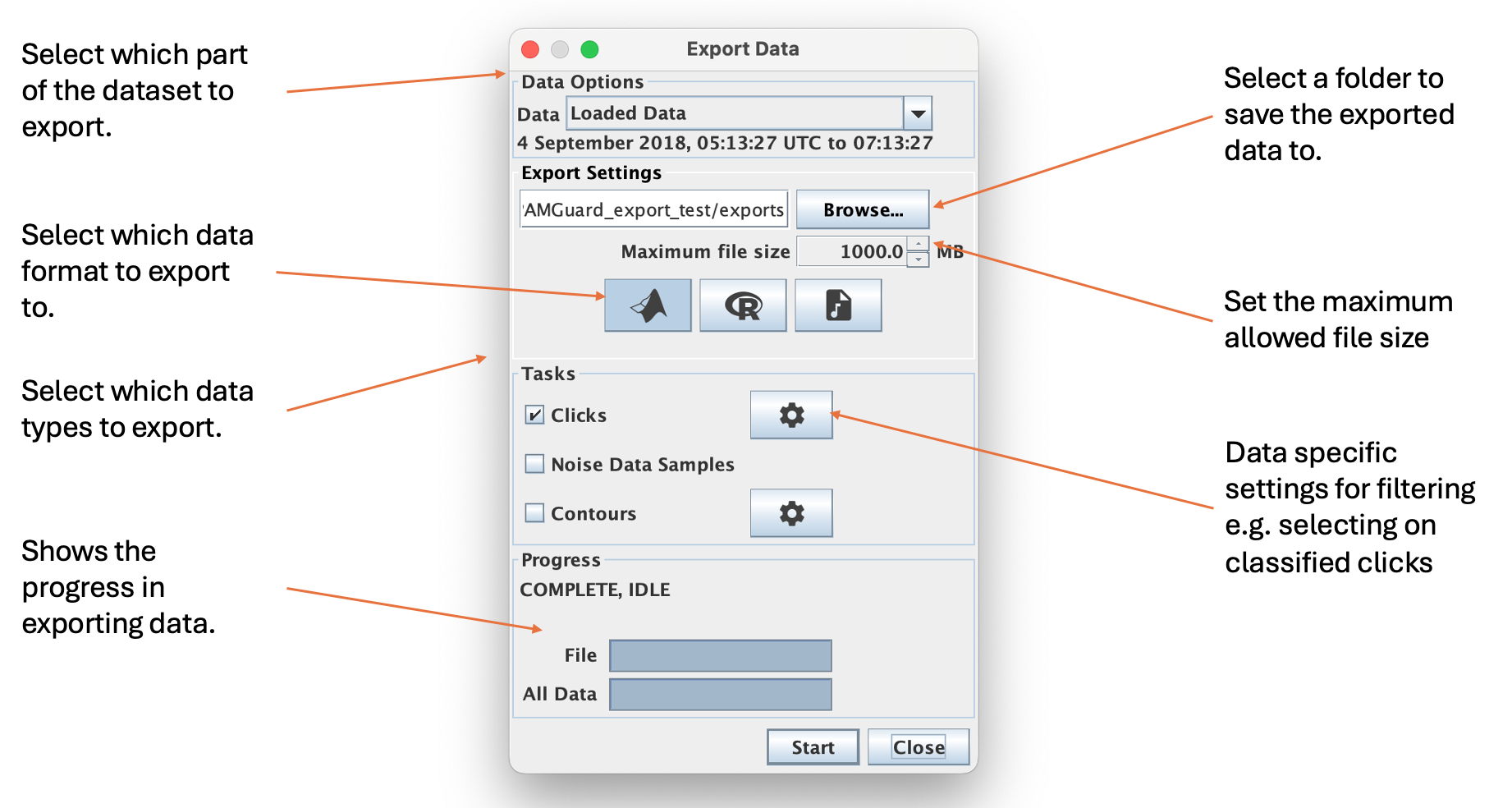
Diagram of the exporter dialog. The dialog allows users to select which part of the dataset to export, how to export it and which type of data to export
The main parts of the dialog are as follows.
Data Options
Select which part of the dataset to export
- Loaded Data : the data currently loaded into memory i.e. usually what you can see in the displays - may be different time oeriods depending on the data type.
- All data : the entire dataset.
- Select data : manually enter a period between two times.
- Specify time chunks : import a csv file with a list of time chunks.
Export Options
Select where to export the data to using Browse… and select the maximum allowed file size using the Maximum file size selector. Select the format by toggling one of the data format buttons. Hover over each button to see more info.
Export Data
Select which data to export. If a data type has a cog icon next to it then it has a data selector. The data selector settings can be used to filter which detections are exported. For example you may wish only to export clicks of a certain type or perhaps deep learning detections with a prediction value above a certain threshold. Each data selector is unique to the type of data. Note that the exporter only exports a sub set of data types - this will expand in future.
Progress
Once Start is selected then the progress bars show progress in exporting the selected data.
Export formats
Currently the exporter has three possible output formats.
MAT files
MAT files are files which can be opened easily in MATLAB and Python. They can store multiple different data formats e.g. tables, arrays, structures. Each PAMGuard detection is saved as a single structure and then the file contains an array of these structures for each data type. The fields within the structure contains the relevant data unique to each data unit. Whilst data units have unique fields depending on their type e.g. a click or a whistle, there are some fields that are shared between almost all data units - an example of a click detection structure is shown below
General fields shared by most data units in PAMGuard
- millis: the unix*1000 start time of the click, whistle, clip etc. in milliseconds; this number can be converted to a date/time with millisecond accuracy.
- date: the start time of the click in MATLAB datenum format. Use datastr(date) to show a time string.
- UID: a unique serial number for the detection. Within a processed dataset no other detection will have this number.
- startSample: The first sample of this detection - often used for finer scale time delay measurements. Samples refers to the number of samples in total the sound card has taken since processing begun or a new file has been created.
- channelMap: The channel map for this detection. One number which represents which channels this detection is from: To get the true channels use the getChannels(channelMap) function.
Unique to clicks
- triggerMap: which channel triggered the detection.
- type: Classification type. Must use database or settings to see what species this refers to.
- duration: Duration of this click detection in samples.
- nChan: Number of channels the detection was made on.
- wave: Waveform data for each channel.
Note that the format of each struct is the same as the format if extracting data using the PAMGuard-MATLAB library.
To open an exported .mat file simply drag it into MATLAB or use the function;
load(/my/path/to/file.mat)To open a .mat file in Python use
import scipy.io
mat = scipy.io.loadmat('/my/path/to/file.mat')
clkstruct = mat['det_20170704_204536_580'] #The name of the struct array within the file
#Extract the third waveform from a click example
nwaves = len(clkstruct[0]) #Number of clicks
thirdwaveform = clkstruct[0, 2]['wave'] #Waveform from third click in samples between -1 and 1. R
Data can be exported to an RData frame. The data are exported as R structs with the same fields as in MATLAB (and PAMBinaries package). To open a an RData frame open RStudio and import the file or use;
load("/my/path/to/file.RData")Wav files
Any detection which contains raw sound data, for example a click, clip or deep learning detection, can be exported as a wav file. When wav files are selected three options are presented for saving files.
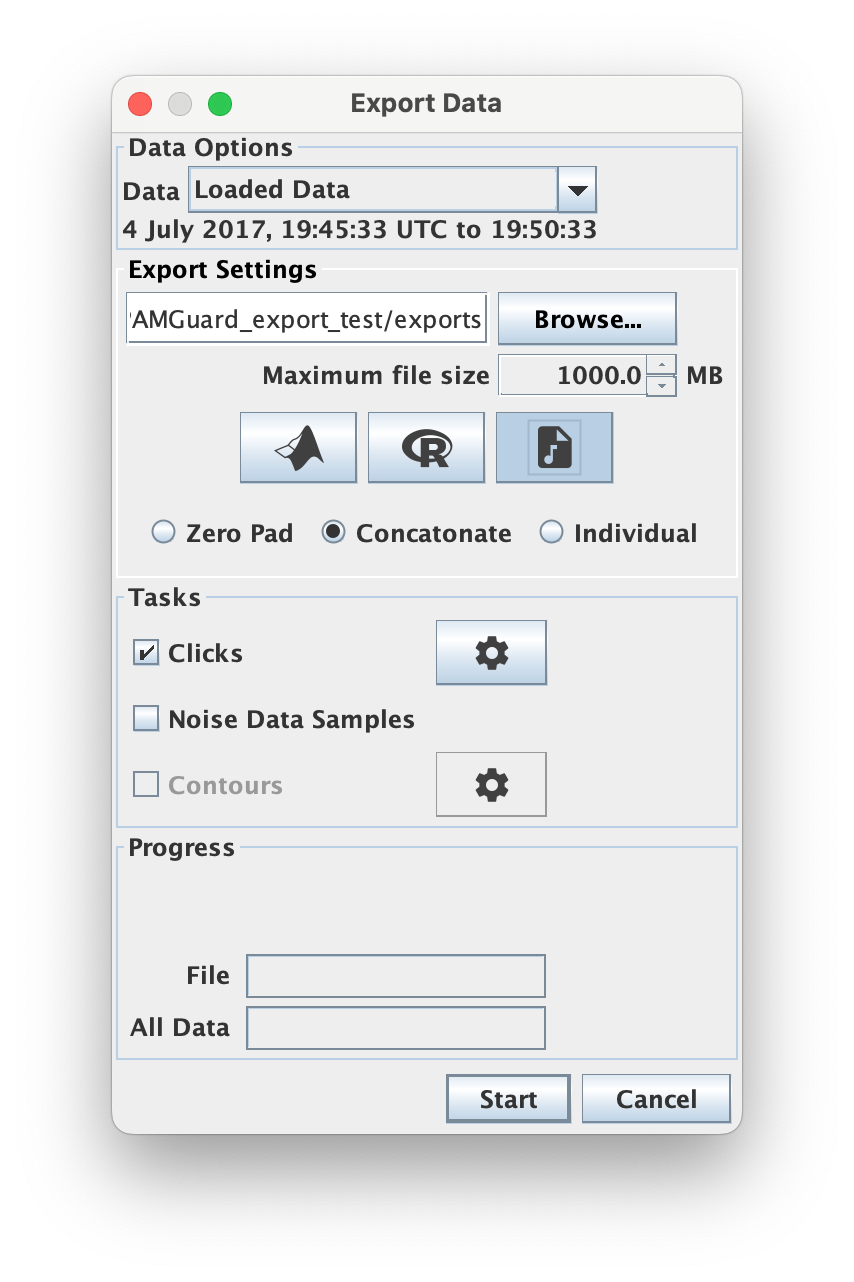
When wav files are selected additional options are presented on how to save the file
- Zero pad : Here detections are saved as wav files with the time in between detections zero padded. The resulting files will be as large as the initial wav files processed to create the data. This can be useful if for example opening the files in another acoustic analysis program.
- Concatenate : The detections are saved to a wav file without any zero padding. This saves storage space but temporal information is lost within the wav file. The sample positions of each detection are saved in a text file along with the wav file so that temporal info is available if needed. This is same format as SoundTrap click detection data.
- Individual : Each detection is saved in it’s own time stamped individual sound file.
After export
Once data are exported, the exported files are not part of PAMGuard’s data management system i.e. PAMGuard has no record they exist and they are not shown in the data model etc. If you export the same data again to the same location, then previous exported files may be overwritten without warning.